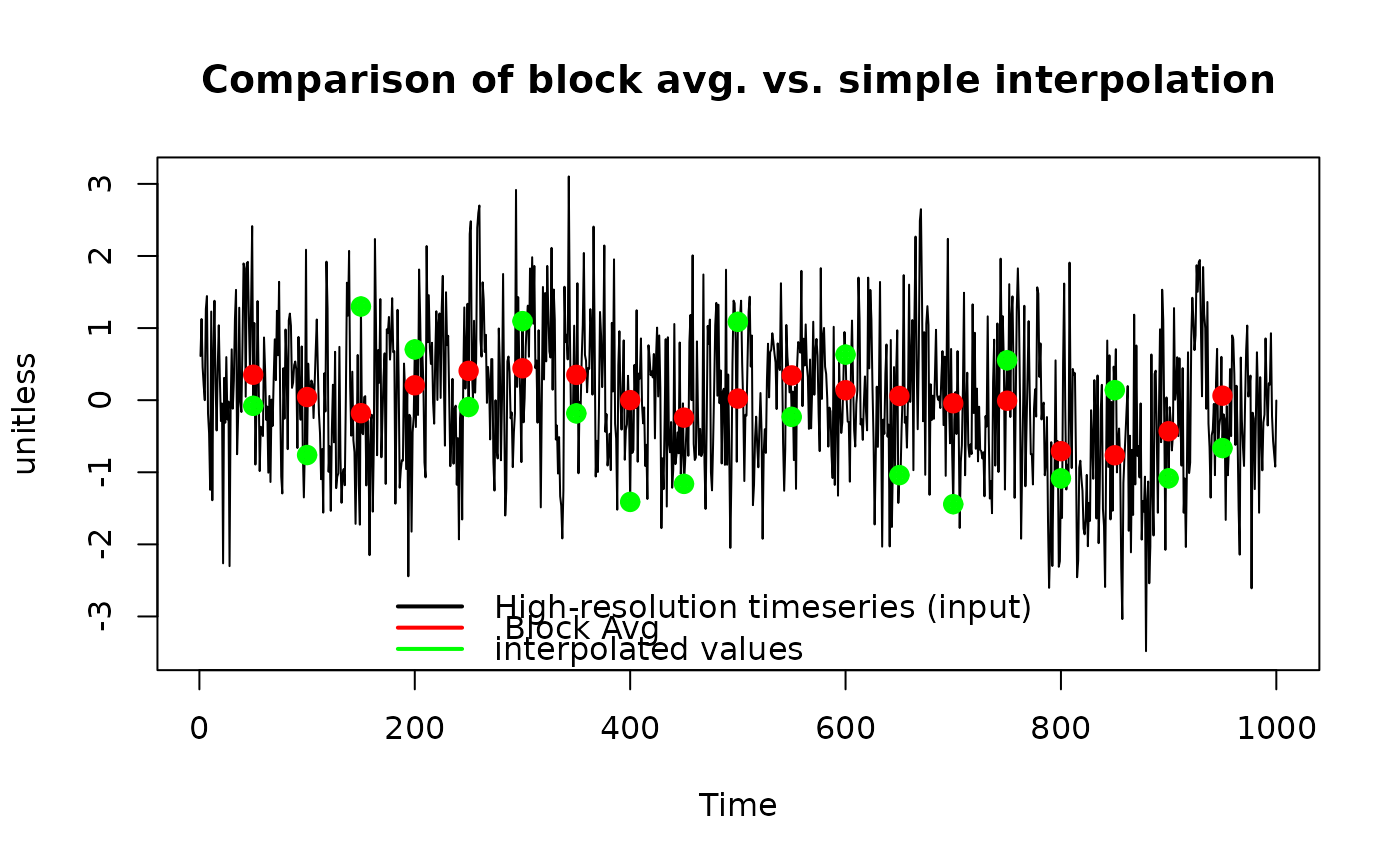
Subsample (downsample) timeseries using block averaging#'
SubsampleTimeseriesBlock.RdResample a equidistant timeseries (e.g. model result) at the 'timepoints' using block averaging. The blocks are divided at 1/2 time between the requested output points. For the first (and last) timepoint, the interval starting mean(diff(timepoints)) before (ending after) are used. Example usage is to downsample a model timeseries to mimick an integrating proxy (e.g. water isotopes that are measured by melting pieces of ice).
Examples
input <- ts(SimPowerlaw(0.5, 1000))
timepoints <- seq(from = 50, to = 950, by = 50)
result <- SubsampleTimeseriesBlock(input, timepoints)
plot(input, main = "Comparison of block avg. vs. simple interpolation",
ylab = "unitless")
points(timepoints, result, pch = 19, col = "red", lwd = 3)
points(approx(time(input), c(input), timepoints), col = "green",
pch = 10, lwd = 3)
legend("bottom", col = c("black", "red", "green"), lwd = 2, bty = "n",
c("High-resolution timeseries (input)", " Block Avg", "interpolated values"))